The role of thin film optics in qPCR and Covid-19 detection
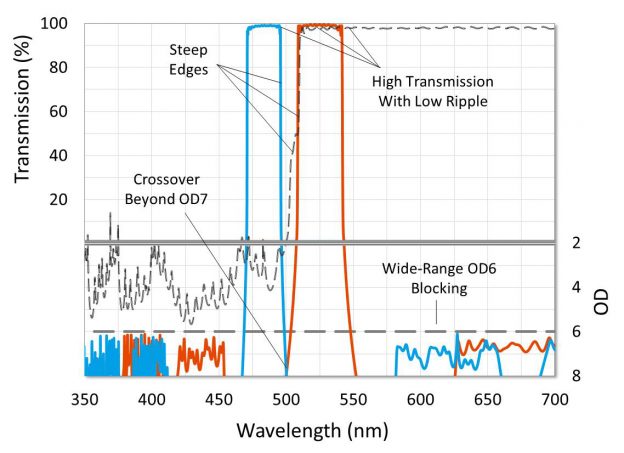
Filter features such as high transmission, steep edges, low ripple and deep blocking are important when testing many samples, says Dr. Rance Fortenberry, director of technology at Alluxa
The current Covid-19 pandemic has highlighted the need for rapid and accurate quantitative analysis of dangerous pathogens, particularly Sars-Cov-2. Fortunately, our ability to determine the structure of new and dangerous viruses has continued to improve since the invention of polymerase chain reaction (PCR), which enables the production of billions of copies of a single DNA sample.
The ‘gold standard’ for DNA detection today is quantitative polymerase chain reaction (qPCR), which combines standard PCR with dye molecules whose fluorescent output increases as the reaction proceeds. This optical monitoring of the PCR reaction in real time – in minutes rather than days – has revolutionised PCR based detection of DNA.
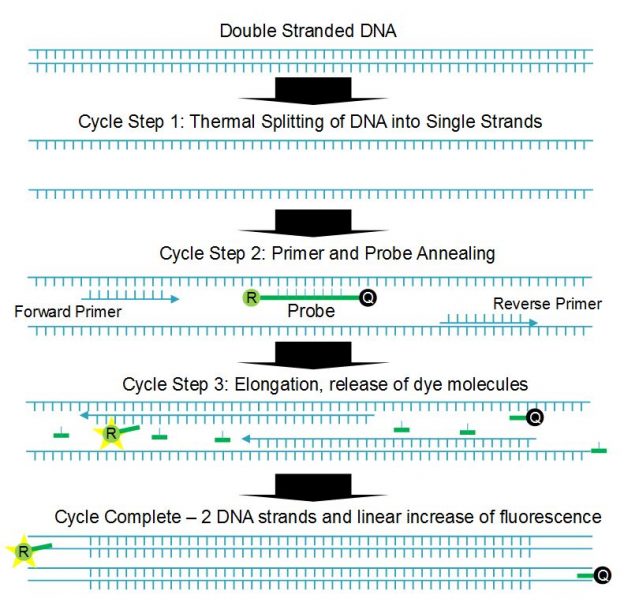
qPCR is based on a multistep thermal cycle (Figure 1). Each cycle, which can take less than a minute, results in a doubling of DNA. Step one separates the sample DNA into single strands by raising the temperature to >90°C, which breaks the hydrogen bonds holding the double stranded DNA together. Step two is an annealing step where the temperature is lowered (55 to 60°C) so that short DNA sequences called primers and probes can attach to the single-stranded DNA. Step three is an extension step where the polymerase enzyme builds the two single strands of DNA into two double strands. Note that although the Sars-Cov-2 virus is RNA based, reverse transcription is used to convert the RNA to complementary DNA before qPCR testing begins.
The short probe and primer DNA sequences are designed to match specific sections of the DNA under analysis, and the probe has the fluorescent dye, or reporter, molecule directly attached along with a quencher molecule. The presence of the quencher molecule prevents fluorescence by allowing only non-radiative energy transfer via a mechanism called Förster resonance energy transfer (FRET). During the extension step the polymerase enzyme breaks up the probe DNA, separates the molecules and removes the quenching effect, leaving the reporter molecule free to fluoresce. Since one reporter molecule is released for each double stranded DNA molecule created, the optical signal scales linearly with the amount of DNA created, or exponentially with cycle number or time.
Typically, multiple sets of primers and probes are created to detect different regions in Sars-Cov-2. Like other coronaviruses, the Sars-Cov-2 virus has four structural proteins known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid). The first three make up the viral envelope surrounding the last, which holds the RNA genome. The United States CDC standard assay uses three N-gene probes, two (N1 & N2) specific to Sars-Cov-2, and one (N3) common to all coronaviruses. A fourth primer and probe set are used to detect human RNAse P (RP) as a check on specimen quality, along with positive and negative controls that check for errors or contamination.
The US CDC assays employ carboxyfluorescein (FAM) as the reporter molecule for all probes, with absorption and emission wavelengths at 495nm and 520nm. In testing, the different probes are in separate wells within a standard 96-well assay plate, with some wells containing probes for N1, others N2, N3, or RP, and others containing positive and negative controls.
When using only a single dye – FAM is the nearly universal choice due to its high brightness – the optics required is particularly simple. A light source (LED, laser, or filtered white light) excites all wells of the assay plate simultaneously with light in the FAM absorption band, and the output fluorescence is passed through a thin film bandpass filter centered on the FAM emission wavelength. The filter output is then imaged onto a CCD camera that quantifies the amount of light emitted by each well as a function of time or cycle number.
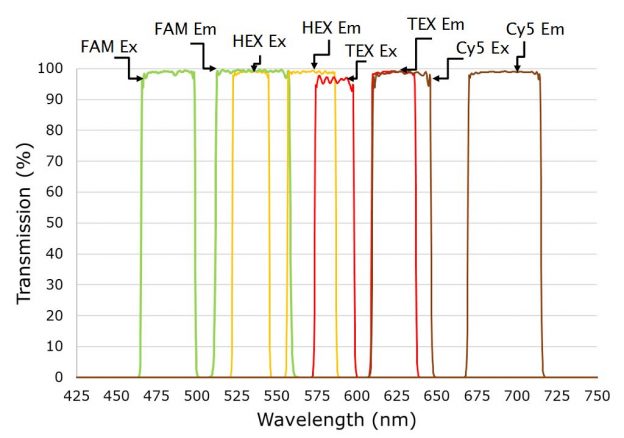
When many samples are to be measured, this relatively simple approach can be improved by using probes with dyes that fluoresce at different wavelengths. Multiplexing more than one probe in a single well can significantly increase the number of samples that can be tested within a given time or budget. However, it is important dye combinations are selected with excitation and emission wavelengths that have minimal overlap, to reduce optical crosstalk, along with quality optical components.
Figure 2b. Four channel setup showing excitation and emission filters multiplexing FAM, HEX, TEX, and Cy5 dyes for Sars-Cov-2 detection
In a single optical channel, higher filter blocking generally reduces unwanted background signal and results in a lower threshold of detection, but in multiplexing systems, optical crosstalk is more critical and can significantly limit testing ability. With available hardware platforms capable of handling four optical channels available today, and new qPCR platforms that multiplex five optical channels starting to arrive, optical filter features such as high transmission, steep edges, low ripple and deep blocking become more important.